Chapter 7 Mixed Model
7.1 introduction
This is summary tutor of linear mixed model. The main reference is https://ourcodingclub.github.io/tutorials/mixed-models/
The data have different hierarchical or grouping characteristics like some patients from different hospitals. In that situation, the stratification analysis is most common methods to figure out exact relationship. However, the lack of power with limit sample size issue came from stratification analysis. So, When those factors all were putted in together, the mixed models were needed. Here, I summary linear mixed models here. In current case summary, We will used covid19 data from 16th countries. The group is country level and the interesting relationship is regarded on individual economic activity and psychological symptoms. The original data are own to Laura, PH.D, Michigan university.
7.2 data step 1
import library
rm(list=ls())
library(tidyverse)
library(htmlTable)
library(readxl)
library(ggrepel)
#library(doMC)
#registerDoMC(31)
library(data.table)
library(DT)
library(plotly)
library(readr)
library(readxl)
library(gridExtra)
library(broom);library(broom.mixed)7.3 load data of Int and Korea
data import of international and korea
#new laura data Dec 2020
#international and korea data
covid = read_csv('data/covid/covid.csv')
kr_d <- read_xlsx('data/covid/kr_data.XLSX', sheet = 1)
cbook <- read_xlsx('data/covid/code_book.xlsx')
# human development index
hdi = read_csv('data/covid/hdi.csv') %>%
setNames(c('rank', 'country', 'y2019', 'country_1'))look up tabel for country name and number
country_lkup <- data.frame(country = c(1, 16, 31, 36,65, 75, 79, 104, 136, 137, 143, 156, 168, 172, 173, 179, 183, 192),
country_c = c(
"USA", "Belarus", "Canada", "China",
"German", "Hong Kong", "Indonesia",
"Malaysia", "Philippines", "Poland",
"Rusia", "Singapore", "Sweden",
"Thailand", "Taiwan", "Turkey",
"Ukraine", "Vietnam"
))overview of codebook
cbook %>% datatable()Korea Data step for value and variable standardization
kr_d1 <- kr_d %>%
select('IDnum' = No, 'age' = SQ2, 'gender' = SQ1, 'Education' = DQ1,
contains('DQ2'), # employment status
'EmoDep' = Q18_2, # for int Q36_2 how much have you feeling
'EmoAnx' = Q18_3, # for int Q36_3
'ChrDep' = Q21_10, # for int Q40_17
'ChrAnx' = Q21_11, # for int Q40_18
'ToDepAnx'= Q22_5, # for int Q41_5,
'dfLossJob' = Q10_10, # for int Q26_9,
'dfReduIcm' = Q10_3, # for int Q26_3,
'dfIncAnx' = Q10_5 # for int Q26_5
) %>%
mutate(country = 200, country_c = 'South Korea') %>%
map_if(is.numeric, ~ifelse(is.na(.x), 0, .x) ) %>%
as.data.frame()international data mutate
international Data step
int_d1 <- covid %>%
mutate('age' = 2020 - Q79) %>%
mutate(IDnum = row_number()) %>%
select(IDnum,
'age',
'gender' = Q10, 'Education' = Q80,
contains('Q94'), # employment status
'EmoDep' = Q36_2,
'EmoAnx' = Q36_3,
'ChrDep' = Q40_17,
'ChrAnx' = Q40_18,
'ToDepAnx'= Q41_5,
'dfLossJob' = Q26_9,
'dfReduIcm' = Q26_3,
'dfIncAnx' = Q26_5,
'country' = Q2
) %>%
left_join(country_lkup, by = c('country')) %>%
select(-Q94_18, -Q94_19)Harmonizing Variable name between Korea and International Data
em_lkup = tibble(
'kr' = c('DQ2_1', 'DQ2_2', 'DQ2_3', 'DQ2_4', 'DQ2_5', 'DQ2_6', 'DQ2_7',
'DQ2_8','DQ2_9','DQ2_10','DQ2_11','DQ2_12','DQ2_13','DQ2_14'),
'int' =c('Q94_9', 'Q94_2', 'Q94_3', 'Q94_4', 'Q94_7', 'Q94_8', 'Q94_5', 'Q94_12', 'Q94_14', 'Q94_15', 'Q94_1', 'Q94_6', 'Q94_16', 'Q94_17')
)
kr_d2<- kr_d1 %>%
plyr::rename(setNames(em_lkup$int, em_lkup$kr)) # change kr variable names to int variable names.
# colnames(kr_d2) == colnames(int_d1) # it returns TRUE value for all col names7.4 data merge
mm <- rbind(int_d1, kr_d2)
nrow(mm)## [1] 19270mm <- mm %>% na.omit(age)
nrow(mm) ## [1] 16942data mutate
krtoint = function(x) {
x = ifelse(x<4, x+4, x)
}
logictozero= function(x){
x = as.numeric(x != 0)
}
mm1 <-mm %>%
filter(age >20 & age <65) %>%
filter(gender %in% c(1, 2)) %>%
filter(!((EmoDep ==-99)|(EmoAnx ==-99)|(dfLossJob==-99)|(Q94_3==-99)|(Q94_4 ==-99)))%>%
mutate (dfLossJob = ifelse(dfLossJob ==10, 1, dfLossJob)) %>%
mutate (Q94_3 = ifelse(!Q94_3 == 0, 1, 0)) %>%
mutate (Q94_4 = ifelse(!Q94_4 == 0, 1, 0))
mm12<- mm1 %>%
mutate(EmoDep =krtoint(EmoDep),
EmoAnx =krtoint(EmoAnx),
ToDepAnx=ifelse(ToDepAnx == 3, 1, 0)) %>%
select(-c('ChrDep', 'ChrAnx', 'dfLossJob','dfReduIcm','dfIncAnx' ))
mm13 <- mm1 %>%
select(c('ChrDep', 'ChrAnx', 'dfLossJob','dfReduIcm','dfIncAnx' )) %>%
lapply(., logictozero) %>%
data.frame()
mm2 <- cbind(mm12, mm13)
mm3 <- mm2 %>%
mutate(EcLoss = ifelse(Q94_3 !=0 | Q94_4!=0, 1, 0)) %>%
mutate(EcLossUnSafe = ifelse(Q94_4!=0, 1, 0)) %>%
mutate(agegp = cut(age, breaks = c(-Inf, (5:13)*5, +Inf))) %>%
mutate(agegp2 = as.numeric(agegp)) %>%
mutate(gender = ifelse(gender ==1, 'Men', 'Women')) %>%
mutate(Edugp = factor(Education, levels = c(1, 2, 3, 4, 5, 6),
labels = c("Middle or less", "High", "Colleage",
"University", "Graduate Sc", "Doctoral or more"))) %>%
mutate(EcLossAll = EcLoss + dfLossJob ) %>%
mutate(TotalDepAnx = ifelse(EmoDep ==7 | EmoAnx ==7, 1, 0))
#mutate(TotalDepAnx= as.numeric((as.numeric(EmoDep ==7) + as.numeric(EmoAnx ==7))) !=0)create basic matrix for visualization (NA as zero)
mm4 <- mm3 %>%
mutate(agegp3 = ifelse(age <=40, '≤40', '>40')) %>%
mutate(agegp3 = factor(agegp3, levels=c('≤40', '>40'))) %>%
filter(Education %in% c(1:6)) %>%
mutate(edugp = ifelse(Education %in% c(1, 2), 1, # high school or less
ifelse(Education %in% c(3),2, # college
ifelse(Education %in% c(4), 3, 4)))) %>% # university (5, 6) Graduate school
mutate(edugp2 = ifelse(Education %in% c(1, 2, 3), 1, 2)) %>%
mutate(EcLossAllgp = ifelse(EcLossAll ==0, 0, 1)) #%>%
#filter(!country %in% c(16, 173))
bm1<-crossing(
'gender'=unique(mm4$gender),
'country_c'=unique(mm4$country_c),
'edugp'=unique(mm4$edugp),
'edugp2'=unique(mm4$edugp2),
'agegp2'=unique(mm4$agegp2),
'agegp3'=unique(mm4$agegp3),
'EcLossAllgp'=unique(mm4$EcLossAllgp),
'TotalDepAnx' = unique(mm4$TotalDepAnx)
)
mm4.0 = mm4 %>%
left_join(hdi %>% filter(!is.na(country_1)) %>%
rename(country_source = country,
country=country_1 ), by = c('country')) %>%
mutate(y2019 = ifelse(country_c =='South Korea', 0.916, y2019))7.5 Data analysis start
7.5.1 job loss due to COVID19, psychological aggravation according to Gender
due to covid vary by country basic static for job loss prevalence across country and genders.
fig1 <- mm4 %>%
group_by(country_c, gender) %>%
count(EcLossAllgp) %>%
mutate(prob = n/sum(n)) %>%
filter(EcLossAllgp == 1 ) %>%
ggplot(aes(x = gender, y = prob, fill = gender)) +
geom_bar(stat = "identity")+
xlab("")+
ylab("Job loss due to COVID19")+
scale_y_continuous(labels = function(x) paste0(x*100, "%"))+
facet_wrap(country_c ~., nrow = 3) +
theme_minimal()+
theme(text = element_text(size=17))+
theme(legend.position = c(.92, .1))
fig2 <- mm4 %>%
group_by(country_c, gender) %>%
count(TotalDepAnx) %>%
mutate(prob = n/sum(n)) %>%
filter(TotalDepAnx == 1 ) %>%
ggplot(aes(x = gender, y = prob, fill = gender)) +
geom_bar(stat = "identity")+
ylab("Psychological aggravation due to COVID19")+
xlab('Gender')+
scale_y_continuous(labels = function(x) paste0(x*100, "%"))+
facet_wrap(country_c ~., nrow = 3) +
theme_minimal()+
theme(text = element_text(size=17))+
theme(legend.position = c(.92, .1))Save total figure
ggsave(arrangeGrob(fig1, fig2, ncol = 1), file ='results/covid/figtotal.png', dpi = 300, width = 10, height =14)
ggsave(arrangeGrob(fig1, fig2, ncol = 1), file ='results/covid/tiff/figtotal.tiff', dpi = 300, width = 10, height =14)
7.5.2 job loss, psychological aggravation according to age group
fig3 <- mm4.0 %>%
#filter(!country %in% c(16, 173)) %>%
group_by(country_c, gender, agegp2) %>%
count(EcLossAllgp) %>%
mutate(prob = n/sum(n)) %>%
filter(EcLossAllgp == 1) %>%
ggplot(aes(x = agegp2, y = prob, color = gender)) +
geom_point(aes(size = n), alpha = 0.2, show.legend = FALSE) +
geom_smooth(method = 'loess', span =0.9, se=FALSE) +
ylab("Job loss due to COVID19")+
scale_y_continuous(labels = function(x) paste0(x*100, "%"))+
theme_minimal() +
xlab("")+
labs(color = "Gender") +
facet_wrap(country_c~., nrow =3)+
theme(legend.position = c(.92, .1))+
theme(text = element_text(size=17))+
scale_x_continuous(breaks = c(2,4,6,8), labels = c(30,40,50,60))
fig4 <- mm4.0 %>%
#filter(!country %in% c(16, 173)) %>%
group_by(country_c, gender, agegp2) %>%
count(TotalDepAnx) %>%
mutate(prob = n/sum(n)) %>%
filter(TotalDepAnx == 1) %>%
ggplot(aes(x = agegp2, y = prob, color = gender)) +
geom_point(aes(size = n), alpha = 0.2, show.legend = FALSE) +
geom_smooth(method = 'loess', span =0.9, se=FALSE) +
ylab("Psychological aggravation due to COVID19")+
scale_y_continuous(labels = function(x) paste0(x*100, "%"))+
theme_minimal() +
xlab("Age (size = number of respondents)") +
labs(color = "Gender") +
facet_wrap(country_c~., nrow =3)+
theme(legend.position = c(.92, .1))+
theme(text = element_text(size=17))+
scale_x_continuous(breaks = c (2,4,6,8), labels = c(30,40,50,60))save figures
#figtotal2 <- grid.arrange(fig3, fig4, ncol = 1)
ggsave(arrangeGrob(fig3, fig4, ncol = 1), file ='results/covid/figtotal2.png', dpi = 300, width = 10, height =14)
ggsave(arrangeGrob(fig3, fig4, ncol = 1), file ='results/covid/tiff/figtotal2.tiff', dpi = 300, width = 10, height =14)
7.6 job loss, psychological aggravation according to Education level
fig5 <- mm4.0 %>%
group_by(edugp, country_c, gender) %>%
count(EcLossAllgp ) %>%
mutate(prob = n/sum(n)) %>%
filter(EcLossAllgp == 1) %>%
ggplot(aes(x = edugp, y = prob, color = gender)) +
geom_smooth(method = 'loess', span =0.9, se = FALSE) +
scale_x_continuous(labels=c("1" = "≤H", "2" = "C",
"3" = "U", "4" = "G")) +
theme_minimal() +
labs(color = "Gender") +
ylab("Job Loss due to COVID19") +
xlab("")+
facet_wrap(country_c ~., nrow = 3)+
theme(text = element_text(size=17))+
theme(legend.position = c(.92, .1))
# mm4 %>%
# count(TotalDepAnx)
fig6 <- mm4.0 %>%
group_by(edugp, country_c, gender) %>%
count(TotalDepAnx ) %>%
mutate(prob = n/sum(n)) %>%
filter(TotalDepAnx == 1 ) %>%
ggplot(aes(x = edugp, y = prob, color = gender)) +
geom_smooth(method = 'loess', span =0.9, se = FALSE) +
scale_x_continuous(labels=c("1" = "≤H", "2" = "C",
"3" = "U", "4" = "G")) +
theme_minimal() +
ylab("Psychological aggravation due to COVID19") +
scale_y_continuous(labels = function(x) paste0(x*100, "%"))+
xlab("H = high school, C = college, U = university, G = graduate ") +
labs(color = "Gender") +
facet_wrap(country_c ~., nrow = 3)+
theme(text = element_text(size=17))+
theme(legend.position = c(.92, .1))save figure
#figtotal3 <- grid.arrange(fig5, fig6, ncol = 1)
ggsave(arrangeGrob(fig5, fig6, ncol = 1), file ='results/covid/figtotal3.png', dpi = 300, width = 10, height =14)
ggsave(arrangeGrob(fig5, fig6, ncol = 1), file ='results/covid/tiff/figtotal3.tiff', dpi = 300, width = 10, height =14)knitr::include_graphics('results/covid/figtotal3.png')
7.6.1 Depression status according job loss status
basic statistic for DepAnx prevalence across country and genders.
7.7 association
(ref: https://cran.r-project.org/web/packages/sjPlot/vignettes/tab_model_estimates.html)
if (!require('sjPlot')) install.packages('sjPlot')
if (!require('sjmisc')) install.packages('sjmisc')
if (!require('sjlabelled')) install.packages('sjlabelled')
library(sjPlot);library(sjmisc);library(sjlabelled)countries <- c("Taiwan","USA", "Belarus", "Canada", "China",
"German", "Hong Kong", "Indonesia",
"Malaysia", "Philippines", "Poland",
"Rusia", "Singapore", "Sweden",
"Thailand", "South Korea", "Turkey",
"Ukraine", "Vietnam")
mm4.1 <- mm4.0 %>%
mutate(TotalDepAnx = factor(TotalDepAnx, levels =c(0,1)),
EcLossAllgp = factor(EcLossAllgp, levels =c(0,1)),
gender = factor(gender, levels =c('Men', 'Women')),
edugp2 = factor(edugp2, levels = c(1,2)),
country_c = factor(country_c, levels= countries))regression model
mod1 = mm4.1 %>%
glm(family = binomial,
formula = TotalDepAnx ~ EcLossAllgp)
mod2 = mm4.1 %>%
glm(family = binomial,
formula = TotalDepAnx ~ EcLossAllgp + age + gender + edugp2)
mod3 = mm4.1 %>%
glm(family = binomial,
formula = TotalDepAnx ~ EcLossAllgp + age + gender + edugp2 +
relevel(as.factor(country_c), ref = 'Taiwan') )tab_model(mod1, mod2, mod3)| TotalDepAnx | TotalDepAnx | TotalDepAnx | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Predictors | Odds Ratios | CI | p | Odds Ratios | CI | p | Odds Ratios | CI | p |
| (Intercept) | 1.34 | 1.29 – 1.39 | <0.001 | 1.37 | 1.19 – 1.58 | <0.001 | 0.88 | 0.72 – 1.09 | 0.257 |
| EcLossAllgp [1] | 1.65 | 1.51 – 1.79 | <0.001 | 1.64 | 1.51 – 1.79 | <0.001 | 1.55 | 1.42 – 1.70 | <0.001 |
| age | 0.99 | 0.99 – 0.99 | <0.001 | 0.99 | 0.99 – 0.99 | <0.001 | |||
| gender [Women] | 1.33 | 1.24 – 1.42 | <0.001 | 1.34 | 1.25 – 1.43 | <0.001 | |||
| edugp2 [2] | 1.34 | 1.24 – 1.43 | <0.001 | 1.28 | 1.19 – 1.39 | <0.001 | |||
|
relevel(country c, ref = “Taiwan”)USA |
1.73 | 1.41 – 2.14 | <0.001 | ||||||
|
relevel(country c, ref = “Taiwan”)Canada |
1.42 | 1.15 – 1.75 | 0.001 | ||||||
|
relevel(country c, ref = “Taiwan”)China |
1.33 | 1.10 – 1.61 | 0.003 | ||||||
|
relevel(country c, ref = “Taiwan”)German |
0.94 | 0.76 – 1.16 | 0.567 | ||||||
|
relevel(country c, ref = “Taiwan”)Hong Kong |
1.22 | 0.97 – 1.53 | 0.093 | ||||||
|
relevel(country c, ref = “Taiwan”)Indonesia |
2.48 | 2.02 – 3.04 | <0.001 | ||||||
|
relevel(country c, ref = “Taiwan”)Malaysia |
1.02 | 0.83 – 1.25 | 0.847 | ||||||
|
relevel(country c, ref = “Taiwan”)Philippines |
1.35 | 1.10 – 1.66 | 0.004 | ||||||
|
relevel(country c, ref = “Taiwan”)Poland |
3.01 | 2.44 – 3.73 | <0.001 | ||||||
|
relevel(country c, ref = “Taiwan”)Singapore |
1.77 | 1.39 – 2.25 | <0.001 | ||||||
|
relevel(country c, ref = “Taiwan”)Sweden |
0.82 | 0.66 – 1.02 | 0.076 | ||||||
|
relevel(country c, ref = “Taiwan”)Thailand |
2.98 | 2.42 – 3.67 | <0.001 | ||||||
|
relevel(country c, ref = “Taiwan”)South Korea |
2.72 | 2.23 – 3.33 | <0.001 | ||||||
|
relevel(country c, ref = “Taiwan”)Turkey |
3.35 | 2.72 – 4.15 | <0.001 | ||||||
|
relevel(country c, ref = “Taiwan”)Ukraine |
1.20 | 0.98 – 1.47 | 0.072 | ||||||
|
relevel(country c, ref = “Taiwan”)Vietnam |
1.89 | 1.54 – 2.33 | <0.001 | ||||||
| Observations | 14243 | 14243 | 14243 | ||||||
| R2 Tjur | 0.009 | 0.022 | 0.065 | ||||||
summay stat of each models.
7.7.1 mixed model
if(!require(lme4)) install.packages('lme4');library(lme4)first mixed model
mm4.2 = mm4.1 %>%
mutate(country_c2 = relevel(as.factor(country_c), ref = 'Taiwan') ) %>%
mutate(TotalDepAnx2 = as.numeric(TotalDepAnx)) %>%
mutate(EcLossAllgp2 = as.numeric(EcLossAllgp)) %>%
mutate(edugp3 = as.numeric(edugp2)) %>%
mutate(age2 = as.numeric(age)) %>%
mutate(gender2 = as.numeric(gender)) mixed.lmer = lmer(TotalDepAnx2 ~ EcLossAllgp2 + age + edugp + gender+
(1|country_c2), data=mm4.2)
summary(mixed.lmer)## Linear mixed model fit by REML ['lmerMod']
## Formula: TotalDepAnx2 ~ EcLossAllgp2 + age + edugp + gender + (1 | country_c2)
## Data: mm4.2
##
## REML criterion at convergence: 19305.2
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -1.8980 -1.0808 0.5415 0.8399 1.5079
##
## Random effects:
## Groups Name Variance Std.Dev.
## country_c2 (Intercept) 0.01083 0.1040
## Residual 0.22546 0.4748
## Number of obs: 14243, groups: country_c2, 17
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 1.4655134 0.0346770 42.262
## EcLossAllgp2 0.0963579 0.0100154 9.621
## age -0.0021919 0.0003372 -6.501
## edugp 0.0228408 0.0040361 5.659
## genderWomen 0.0653560 0.0080437 8.125
##
## Correlation of Fixed Effects:
## (Intr) EcLsA2 age edugp
## EcLssAllgp2 -0.409
## age -0.459 0.090
## edugp -0.357 0.064 0.057
## genderWomen -0.164 0.027 0.103 -0.007#qqnorm(resid(mixed.lmer))mixed.lmer2 = lmer(TotalDepAnx2 ~ EcLossAllgp + age2 +
(1|edugp3)+ (1|country_c2/gender2), data=mm4.2)
summary(mixed.lmer2)## Linear mixed model fit by REML ['lmerMod']
## Formula:
## TotalDepAnx2 ~ EcLossAllgp + age2 + (1 | edugp3) + (1 | country_c2/gender2)
## Data: mm4.2
##
## REML criterion at convergence: 19288.9
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -1.9338 -1.0857 0.5291 0.8523 1.5039
##
## Random effects:
## Groups Name Variance Std.Dev.
## gender2:country_c2 (Intercept) 0.003000 0.05477
## country_c2 (Intercept) 0.008937 0.09453
## edugp3 (Intercept) 0.001716 0.04142
## Residual 0.224888 0.47422
## Number of obs: 14243, groups:
## gender2:country_c2, 34; country_c2, 17; edugp3, 2
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 1.646672 0.041157 40.010
## EcLossAllgp1 0.095570 0.010010 9.548
## age2 -0.002198 0.000338 -6.503
##
## Correlation of Fixed Effects:
## (Intr) EcLsA1
## EcLssAllgp1 -0.084
## age2 -0.343 0.091#qqnorm(resid(mixed.lmer2))mixed.lmer3 = lmer(TotalDepAnx2 ~ EcLossAllgp + age2 + edugp2+
(1+TotalDepAnx2|country_c2/gender2), data=mm4.2)
summary(mixed.lmer3)## Linear mixed model fit by REML ['lmerMod']
## Formula: TotalDepAnx2 ~ EcLossAllgp + age2 + edugp2 + (1 + TotalDepAnx2 |
## country_c2/gender2)
## Data: mm4.2
##
## REML criterion at convergence: -394555.3
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -3.702e-06 -2.610e-07 1.009e-07 3.772e-07 2.663e-06
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## gender2:country_c2 (Intercept) 8.457e-06 2.908e-03
## TotalDepAnx2 2.315e-03 4.812e-02 -0.86
## country_c2 (Intercept) 5.900e-05 7.681e-03
## TotalDepAnx2 4.791e-05 6.922e-03 0.68
## Residual 4.745e-14 2.178e-07
## Number of obs: 14243, groups: gender2:country_c2, 34; country_c2, 17
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 1.431e-02 1.932e-03 7.408
## EcLossAllgp1 1.691e-14 4.623e-09 0.000
## age2 1.732e-15 1.562e-10 0.000
## edugp22 1.311e-14 4.080e-09 0.000
##
## Correlation of Fixed Effects:
## (Intr) EcLsA1 age2
## EcLssAllgp1 0.000
## age2 0.000 0.085
## edugp22 0.000 0.067 0.069
## optimizer (nloptwrap) convergence code: 0 (OK)
## unable to evaluate scaled gradient
## Model failed to converge: degenerate Hessian with 1 negative eigenvalues#qqnorm(resid(mixed.lmer2))make region variable, humain devepin index and .. stratification anaysis. (Asia vs non-Asia or …) deveolping index ..
7.7.2 mixed model with human develop index stratification
agu1 <- read_csv('data/covid/covid_agu1.csv')
lab_mm <- mm4.0 %>%
left_join(agu1, by = 'country_c') %>%
mutate(inc_aug = c_case_agu1 / population *100000,
dth_aug = c_death_agu1/ population *100000) %>%
group_by(country_c, gender) %>%
summarize(hdi = mean(y2019),
Psycho = mean(TotalDepAnx ==1),
ecl = mean(EcLossAll !=0),
inc = mean(inc_aug))
lab_mm%>%
ggplot(aes(x = hdi, y = ecl, color = inc)) +
geom_point() +
theme_classic()+
xlab("Human Devlopment Index") +
#ylab("prevalance of Psychological Symptoms") +
ylab("prevalance of Economic Loss due to COVID19") +
#ylim(c(-0.1, 1)) + #xlim(c(2, 4)) +
geom_label_repel(aes(label = country_c),
fill = NA, # 투명하게 해서 겹쳐도 보이게
alpha =1, size = 3, # 작게
box.padding = 0.4, # 분별해서
segment.size =0.1, # 선
force = 2) + # 이것은 무엇일까요?
theme_minimal() +
geom_smooth(method = 'lm', formula = y ~ poly(x,2), se=FALSE) +
#geom_smooth( se=FALSE) +
facet_wrap(gender~.) +
guides(color = "none")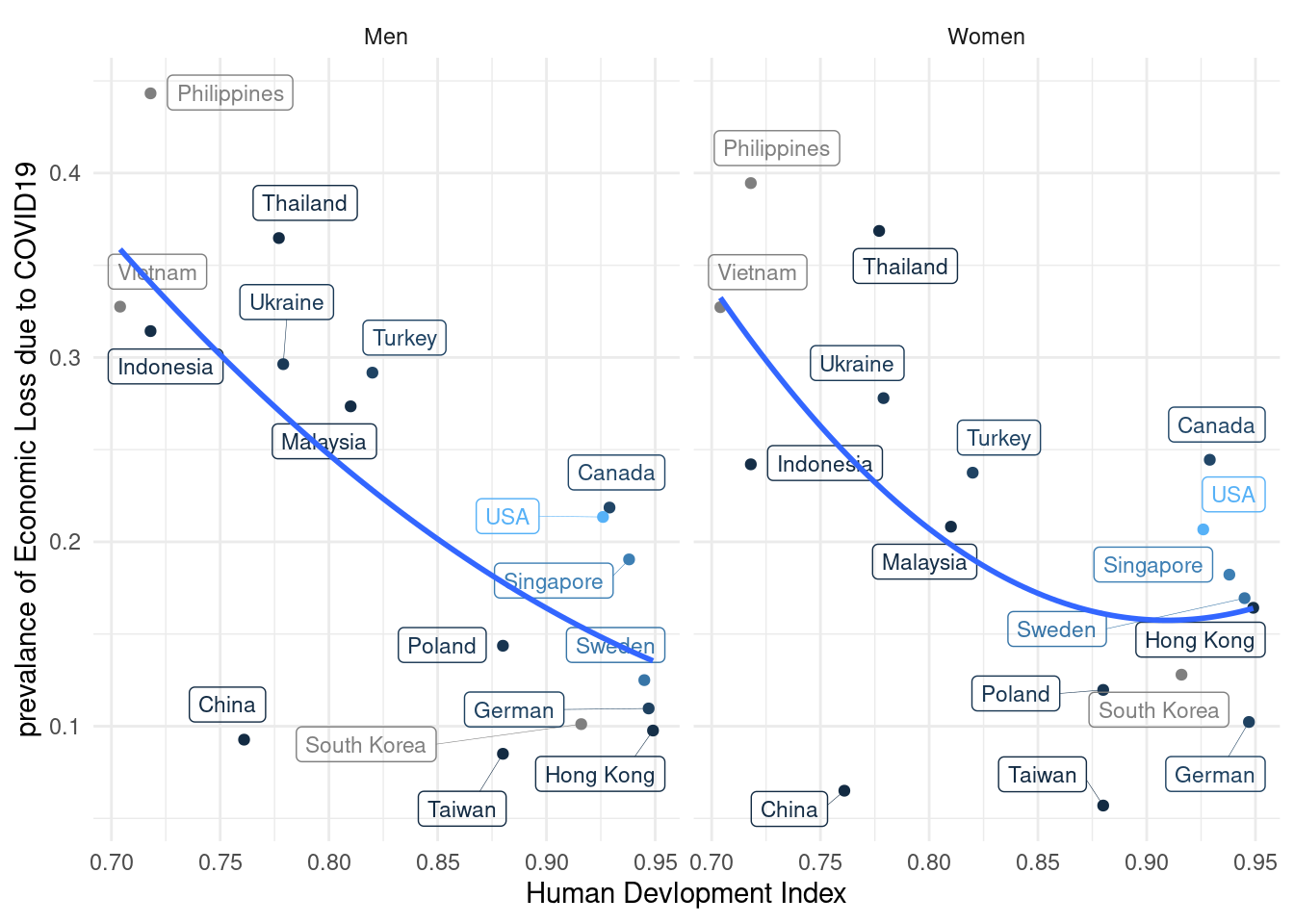
mixed.lmer4 = lmer(TotalDepAnx2 ~ EcLossAllgp2 + age2 +
(1|country_c), data=mm4.2 %>% filter(edugp2 ==2))
summary(mixed.lmer4)## Linear mixed model fit by REML ['lmerMod']
## Formula: TotalDepAnx2 ~ EcLossAllgp2 + age2 + (1 | country_c)
## Data: mm4.2 %>% filter(edugp2 == 2)
##
## REML criterion at convergence: 12714
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -1.8649 -1.1252 0.5519 0.8157 1.4511
##
## Random effects:
## Groups Name Variance Std.Dev.
## country_c (Intercept) 0.01124 0.1060
## Residual 0.22338 0.4726
## Number of obs: 9436, groups: country_c, 17
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 1.6157913 0.0351975 45.906
## EcLossAllgp2 0.0874493 0.0123267 7.094
## age2 -0.0028828 0.0004268 -6.754
##
## Correlation of Fixed Effects:
## (Intr) EcLsA2
## EcLssAllgp2 -0.455
## age2 -0.518 0.072#qqnorm(resid(mixed.lmer2))mod3 = mm4 %>% glm(family = binomial, formula = TotalDepAnx ~ EcLossAllgp + age + gender + edugp2 + relevel(as.factor(country_c), ref = ‘Taiwan’) )
mixed.lmer1 = lmer(TotalDepAnx2 ~ EcLossAllgp2 + age + edugp + gender+
(1|country_c2), data=mm4.2 %>% filter(y2019 >0.9))
summary(mixed.lmer1)## Linear mixed model fit by REML ['lmerMod']
## Formula: TotalDepAnx2 ~ EcLossAllgp2 + age + edugp + gender + (1 | country_c2)
## Data: mm4.2 %>% filter(y2019 > 0.9)
##
## REML criterion at convergence: 6860.8
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -1.8989 -1.0078 0.5255 0.8818 1.5447
##
## Random effects:
## Groups Name Variance Std.Dev.
## country_c2 (Intercept) 0.01017 0.1008
## Residual 0.23064 0.4802
## Number of obs: 4962, groups: country_c2, 7
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 1.411453 0.055829 25.282
## EcLossAllgp2 0.127543 0.018872 6.758
## age -0.002507 0.000560 -4.477
## edugp 0.014514 0.006520 2.226
## genderWomen 0.102722 0.013814 7.436
##
## Correlation of Fixed Effects:
## (Intr) EcLsA2 age edugp
## EcLssAllgp2 -0.435
## age -0.513 0.107
## edugp -0.343 -0.006 0.090
## genderWomen -0.194 -0.015 0.117 0.064mixed.lmer2 = lmer(TotalDepAnx2 ~ EcLossAllgp2 + age + edugp + gender+
(1|country_c2), data=mm4.2 %>% filter(y2019 <=0.9 & y2019 >=0.8))
mixed.lmer3 = lmer(TotalDepAnx2 ~ EcLossAllgp2 + age + edugp + gender+
(1|country_c2), data=mm4.2 %>% filter(y2019 <=0.8))
hdi0.9 = summary(mixed.lmer1)$coefficient[2, 1] %>% exp()
hdi0.8 = summary(mixed.lmer2)$coefficient[2, 1] %>% exp()
hdi0.7 = summary(mixed.lmer3)$coefficient[2, 1] %>% exp()
hdi0.9ci = confint(mixed.lmer1)[4, ] %>% exp()
hdi0.8ci = confint(mixed.lmer2)[4, ] %>% exp()
hdi0.7ci = confint(mixed.lmer3)[4, ] %>% exp()
hdi0.9ci## 2.5 % 97.5 %
## 1.094813 1.178854tibble(c(hdi0.9, hdi0.9ci),
c(hdi0.8, hdi0.8ci),
c(hdi0.7, hdi0.7ci)) %>%
setNames(c('OR', 'LL', 'UL')) %>%
mutate(HDI = c('>0.9', '>0.8', '<0.8')) %>%
select(HDI, OR, LL, UL)## # A tibble: 3 × 4
## HDI OR LL UL
## <chr> <dbl> <dbl> <dbl>
## 1 >0.9 1.14 1.09 1.09
## 2 >0.8 1.09 1.04 1.06
## 3 <0.8 1.18 1.13 1.12